 When Dr. Gavin Naylor and his team started a genetic survey of existing shark and ray species, they didn’t expect the results of their project to make international news. Their recent paper (which, at over 250 pages and complete with more than 100 figures, is nothing short of epic), however, is too striking to ignore. The results indicate that there may be as many as 79 previously unrecognized cryptic species of sharks and rays.
When Dr. Gavin Naylor and his team started a genetic survey of existing shark and ray species, they didn’t expect the results of their project to make international news. Their recent paper (which, at over 250 pages and complete with more than 100 figures, is nothing short of epic), however, is too striking to ignore. The results indicate that there may be as many as 79 previously unrecognized cryptic species of sharks and rays.
A cryptic species is defined as a group that looks almost exactly like another, and may even live in the same region, but is genetically distinct. We’ve known that cryptic species of sharks and rays exist for some time, such as manta rays and scalloped hammerhead sharks, but 79 is a lot; as of the paper’s publication, only 1,221 species of sharks and rays were recognized.
According to Dr. Naylor,
“Organisms become genetically differentiated over time through the cumulative effects of mutation and recombination mediated via drift and selection. When they differentiate in isolation they eventually become so different from the parental stock from which they were derived that they can no longer produce fertile offspring when crossed with them. Some biologists use the point of reproductive inviability as the point at which new species should be recognized….. For practical purposes we recognize “new species” as being genetically or morphologically distinctive from previously recognized forms.”
The study’s methods, though enormous in scope, were relatively basic. According to Dr. Naylor, the study utilized a technique very familiar to geneticists: “standard DNA extraction, PCR, Sanger sequencing, alignment and analysis of a protein coding mitochondrial gene”. To achieve the goals of understanding both evolutionary relationships of sharks and rays and parasite host specificity ( where certain parasites associated only with one species), Dr. Naylor and his team obtained and analyzed samples from as many species as they could. The numbers are impressive- 56 of 57 known families of elasmobranchs were represented among the 4,283 samples from 305 species of sharks and 269 species of batoids. In other words, this study included approximately half of all known elasmobranch species, including many that had never been analyzed genetically before. Since 1986, when the project began, samples have been obtained in more than 50 countries, mostly through the team’s own field work!
For readers unfamiliar with sequencing and alignment of mitochondrial genes, Dr. Naylor explains the methods as follows:
“Molecular tools are useful for ascertaining the degree of differentiation among DNA sequences. Essentially, we align a string of nucleotides for a particular gene of interest and count the proportion of sites that differ between forms. Individuals within species are generally quite similar in sequence while individuals from other species are noticeably different. However, the genes that are generally used for monitoring such differentiation [mitochondrial genes] , are usually not associated with the mechanistic/reproductive basis for the species distinction. They are merely passively evolving markers whose differentiation is more a reflection of the passage of time than any sort of reproductive isolation. In general reproductive isolation tends to be correlated with the passage of time. But not always. “
After the mitochondrial genes were sequenced, the rest is a simple matter of computer alignment (which took several days), and the overall resulting tree is shown below. Don’t worry if you can’t see details clearly- each number along the perimeter represents its own figure. Yes, you read that correctly, there is so much information in this tree that it needs to be broken down into 77 different figures in order to read it. I’ll say it again, this paper is epic.
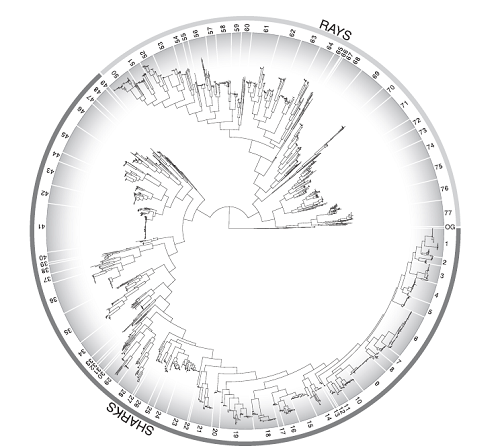
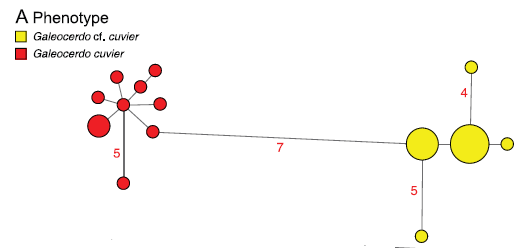
If several samples from different individuals of what’s thought to be one species don’t cluster together, it indicates that they may actually be two different species. This occurred 79 times in this study- 38 times for sharks and 41 for rays. A figure called a parsimony haplotype network can corroborate this when all the samples for one species are plotted together. For example, with tiger sharks, there is absolutely no overlap of haploptype between two different populations of these animal- the DNA sequences found in the mitochondrial genes of the Australian/Pacific ocean subpopulation of tigers is totally different from the Atlantic/Gulf of Mexico subpopulation (see figure below).
So, are there really 79 new species of sharks and rays? Not necessarily, according to Dr. Naylor.
“There are circumstances where we find individuals that are quite divergent genetically and yet capable of interbreeding to produce fertile offspring. On the other hand, there are species that are biologically and functionally distinct but may have very similar nucleotide sequences. In generally we feel more comfortable asserting that two species are distinct when we find genetic divergence in association with morphological differentiation. Morphological differences are assumed to be a reflection of more widespread modifications in organismal architecture and therefore more reliable than a single marker gene.”
Yet, if there are in fact 79 previously unrecognized cryptic species, it isn’t good news for conservation. Scalloped hammerheads are already considered Endangered by the IUCN Shark Specialist Group. If they are actually two different species, it means that each is even more endangered (scientists haven’t discovered a new population of many individual sharks, just that the existing sharks may be be divided into more species than previously thought). Dr. Naylor explains:
“In most cases if a species is considered endangered and then we find out that the species is in fact two different species then, of course, it’s potentially very serious and we quickly need to find out the extent of the ranges, depths and habitats that are associated with each of the species to best manage them… In cases where species that were once thought to be wide ranging, turn out to be a collection of genetically distinct species, each with restricted ranges, the prognosis for survival could be dire. What might have been regarded as extirpation of a local population of a wide-ranging species, might actually be the extinction of an undocumented species.”
Not all the news is bad, though. This study also identified some examples of the opposite situation: that in some cases, what was thought to be two different and endangered populations are actually one population that’s less endangered. The best example of this is the Galapagos shark and the dusky shark (the latter of which is so depleted that the United States shark fisheries management plan estimates it will take slightly for them to recover than there has been a United States).
“The Galapagos Shark (Carcharhinus galapagensis), for example, was first described taxonomically in 1905. It occurs only on tropical oceanic islands such as the Galapagos. The very similar looking Dusky shark (Carcharhinus obscurus), described in 1818, is distinguished from the Galapagos shark primarily by its geographic distribution. It does not occur on oceanic islands, but rather is restricted to continental coastlines. Our genetic data, however, suggest that Galapagos and Dusky sharks are in fact, one and the same. Indeed, recent tagging data compiled by graduate student Corey Eddie at the University of Massachusetts corroborates this suggestion. There are cases where Galapagos sharks that have been tagged on offshore islands (Bermuda) and then show up on continental coastlines…. If Galapagos and Dusky sharks are one and the same species, then their prospects for survival are perhaps not as dire as previously feared,” Dr. Naylor said
Follow-up studies which focus on attributes like morphology, behavior, and life history can help clarify which of these 79 are really previously unrecognized species which require new conservation and management plans. In the meantime, congratulations are due to Dr. Naylor and his team for a great study that will change how we think about the evolution and speciation of elasmobranchs.

Update: The paper is now freely available online. Please click here to download it.
I am so happy this study stood up to expert scrutiny! Also, I want the genome figure on a tshirt!
Dude! No link to the paper?
I hope their data is all available through an open access service like Dryad, because we can ask some seriously interesting questions about shark evolution and demographic history with this set.